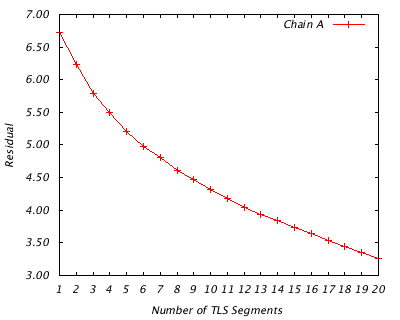
For each protein chain, this analysis show the optimal division into 1 TLS group, 2 TLS groups, 3 TLS groups, etc, up to 20 groups. It goes on to analyze the implied rigid body translational and rotational motion of each group, as well as its quality of fit to the refined atomic displacement parameters (B-factors). If you have multiple chains in your structure, they are treated independently.