 MSMS + Raster3D
MSMS + Raster3DThis figure illustrates using Raster3D to draw a slice through a molecular surface, revealing interior cavities and channels.
Full size image:
PNG format (280 kByte)
JPEG format (49 kByte)
 MSMS + Raster3D
MSMS + Raster3DThis figure illustrates using Raster3D to draw a slice through a molecular surface, revealing interior cavities and channels.
Full size image: |
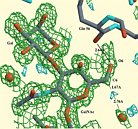 XtalView + Raster3D
XtalView + Raster3DYou can render the canvas window of an XtalView map-fitting session as a Raster3D image via a simple pull-down menu.
Full size image: |
 XtalView + Raster3D
XtalView + Raster3DBoth XtalView and Raster3D can now represent thermal ellipsoids. This view of ellipsoids + density from the 1.25Å refinement of CTB was composed by merging files from Xfit and rastep, and then labeled using ImageMagick.
Full size image: |
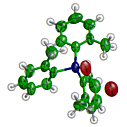 ORTEX + Raster3D
ORTEX + Raster3DPatrick McArdle has adapted the Raster3D components rastep and render for use with the small-molecule visualization tool ORTEX.
Full size image: |
|
Position of Bee Venom Phospholipase A2 at the Membrane Surface Using a Novel Electron Paramagnetic Resonance Technique (image courtesy of Ellie Adman).
Full size image: |
|
Full size image: |
|
Full size image: |
|
Full size image: |
|
Full size image: |
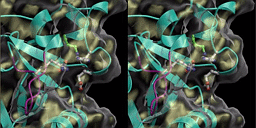 Raster3D can render transparent surfaces.
The stereo pair shown here was built up by merging a
description of protein secondary structure from Molscript
and the corresponding molecular surface as caculated by
Anthony Nicholl's program GRASP.
Raster3D can render transparent surfaces.
The stereo pair shown here was built up by merging a
description of protein secondary structure from Molscript
and the corresponding molecular surface as caculated by
Anthony Nicholl's program GRASP.
Full size image: |
 An MPEG movie:
Clemens Vonrhein and Gerd Schlauderer (University of Freiburg)
have used Molscript and Raster3D to prepare an animation of adenylate
kinase moving between the "open" and "closed" conformations.
More information on
"Adenylate Kinase - the movie"
is available from the authors via WWW.
This work was described in Structure 3: 483-490 (1995).
An MPEG movie:
Clemens Vonrhein and Gerd Schlauderer (University of Freiburg)
have used Molscript and Raster3D to prepare an animation of adenylate
kinase moving between the "open" and "closed" conformations.
More information on
"Adenylate Kinase - the movie"
is available from the authors via WWW.
This work was described in Structure 3: 483-490 (1995).
An
MPEG version of the movie is available here.
|