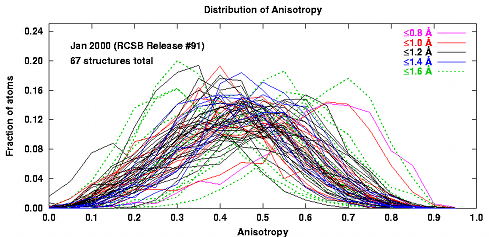
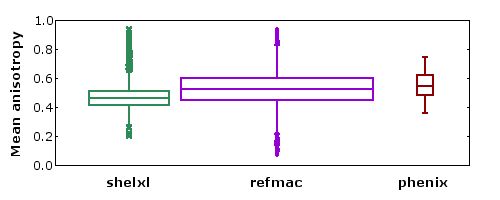
- The 1000+ structures refined using shelxl are mostly at atomic resolution.
- The 5000+ structures refined using refmac are mostly TLS models refined at moderate resolution.
- It is not clear whether the slight difference in the mean value of A is due to the program, the resolution, or the use of TLS models.
- There are outliers both at high and low extremes of mean A.
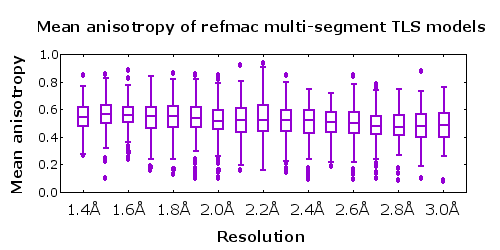
- Structural models at all resolutions exhibit roughly the same anisotropy A (lower figure).
In my experience, full anisotropic refinement at resolutions worse than 1.40Å is improved by adjusting the restraint weights to force the resulting distribution of anisotropy nearer to that seen for the higher resolution structures. This can be confirmed by following the Rfree residual. However, at these resolutions it is a good idea to try a multi-group TLS model as an alternative. The TLSMD server can help you select a multi-group TLS model.