Structure determinations and Structure-based Inhibitor Design

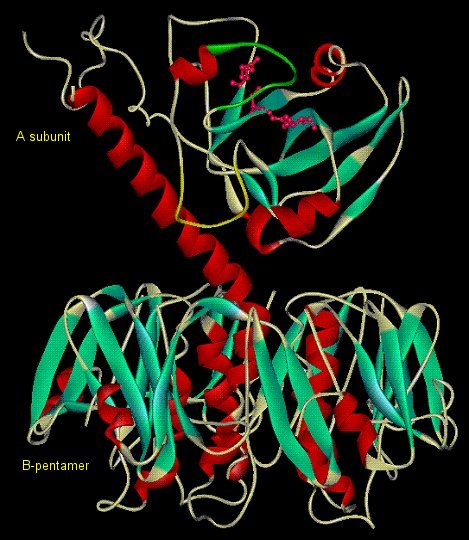
Cholera Toxin - Causative agent of cholera, secreted by Vibrio
cholerae while in the small intestine. Heterohexameric (AB5) protein toxin
B5 allows entry into epithelial cells by docking on GM1 gangliosides. Enzymatically
active A catalyzes ADP-ribosylation of GSα.
(Claire O'Neal)

Close-up of the active site of the A subunit of cholera toxin.
In green: the active site loop, in yellow the so-called activation loop.
(Claire O'Neal)
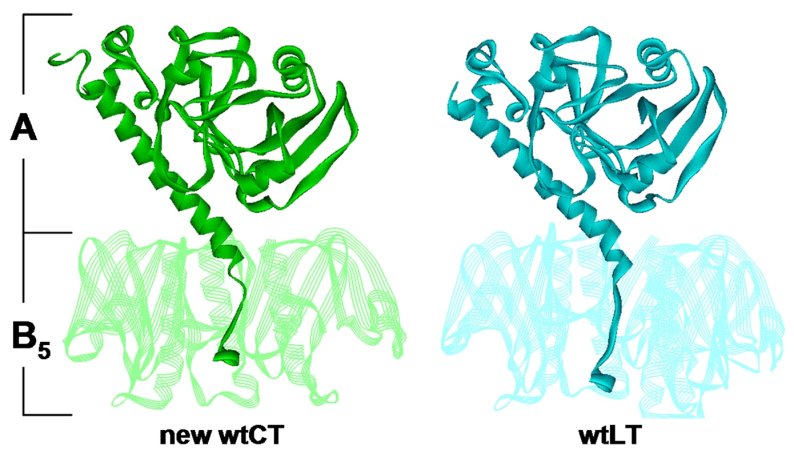
Comparison of CT and LT holotoxin in structures in similar
orientations. Wild-type CT form 1 molecule 1 is on the left, and the 1.9Å
structure of wild-type LT is on the right. B5 is shown in semitransparent
ribbons with from subunits removed to reveal the A2 tail detail.
(Claire O'Neal)
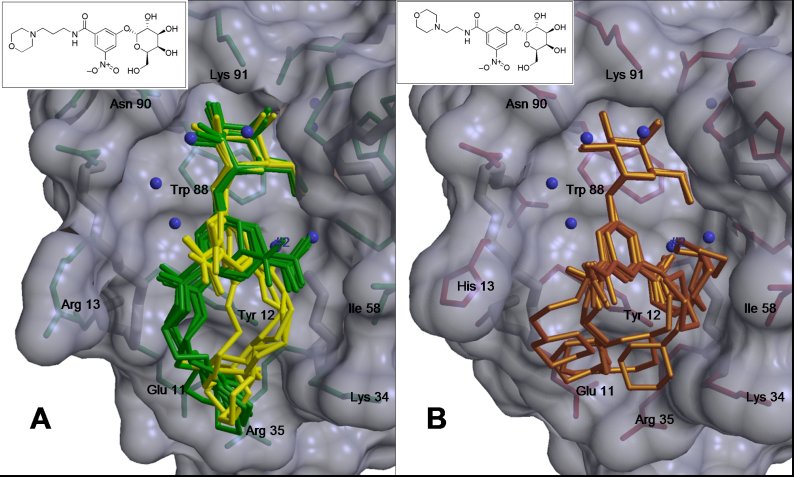
Previously determined co-crystal structures of compounds 1 (A, yellow and
green) and 2 (B, gold) with LTB5 and CTB5 respectively.
(Ethan Merritt)
Section I 1.2 Trypanosomal Glycolytic Enzymes
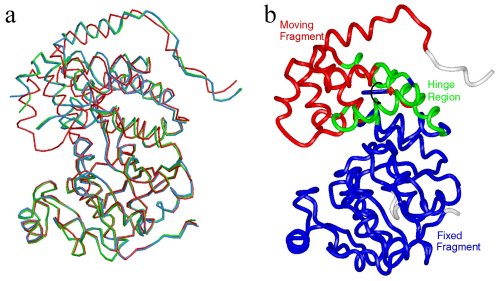
Conformational changes of Leishmania major Glycerol-3-phosphate
Dehydrogenase (LmGPDH). (a) Superposition of apo, binary (E.NADH)
and ternary (E.NAD+-DJAP) LmGPDH structures.
They are colored blue, green and red, respectively. (b)Ca
atom trace of the ternary LmGPDH structure is colored according to
the results of the program Dyndom used for domain motion analysis.
(Jungwoo Choe)

Sigma A weighted Fo - Fc map contoured
at 2.4σ level before model building shown with the final model of the
adduct.
(Jungwoo Choe)
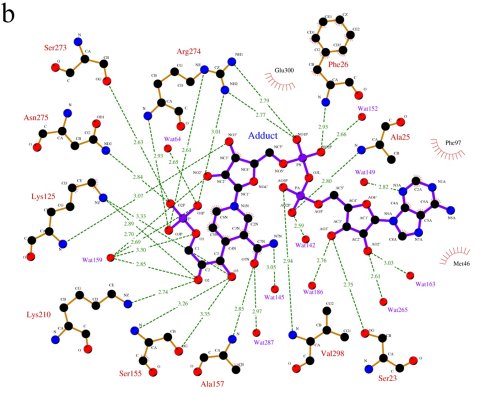
A LIGPLOT representation of the active site, showing
the interactions between LmGPDH and the adduct molecule.
(Jungwoo Choe)
Trypanosomal Peroxin 5
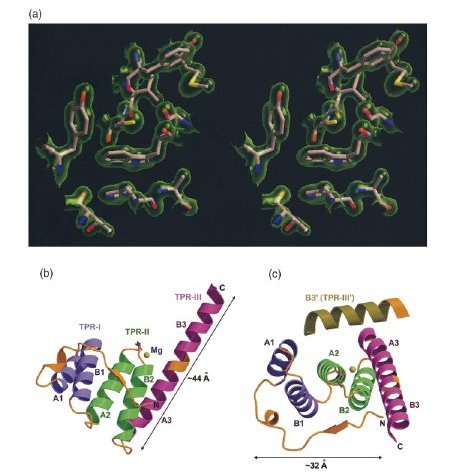
Electron density map, the overall structure, and the Mg2+ binding site of T. brucei PEX5-T3.
(a) Stereo view of a typical region in the 2Fo-
Fc map contoured a 1σ above the mean with the refined
model of the protein superimposed in the map.
(b) The overall structure of PEX5-T3. The N-terminal coil region and all the
loop regions in the structure are shown in gold, while the three TPR motifs
are depicted in purple, green and fuschia, respectively. The A and B helices
in each TPR as well as the Mg ion contacting residue E397 have been labeled.
The third TPR motif does not conform to the standard two-helix TPR motif,
but exists as a continuous long helix. The putative loop region connecting
the two helices in TPR-III is depicted in gold.
(Abhinav Kumar)
Section I 1.3 Human Tyrosyl-DNA- Phosphodiesterase (Tdp1)

Electron density associated with a cluster of tyrosine
residues in the C-terminal domain of Tdp1.
(Doug Davies)
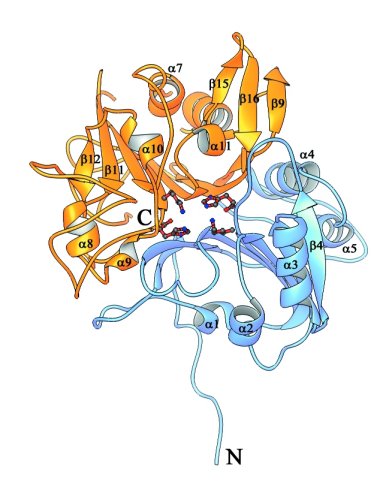
Ribbon representations of the structure of Human Tdp1.
Stereoview of Tdp1 colored to show the two-domain structure of the protein.
The N-terminal domain of Tdp1 (residues 149-350) is colored blue, and the
C-terminal domain (residues 351-608) is colored yellow. The active site residues
His263, Lys265, His493, and Lys495 are depicted as red ball and stick structures.
(Doug Davies)
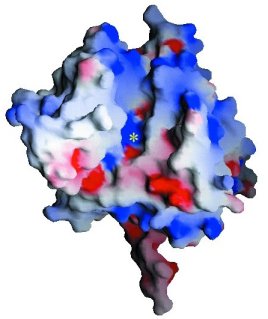
Stereoview of the electrostatic potential surface of
Tdp1. Electrostatic surfaces are colored between -10kT (red) and +10kT (blue).
View parallel to the pseudo-2-fold axis of symmetry between the two domains
of Tdp1. The location of the active site is marked with a yellow asterisk.
(Doug Davies)
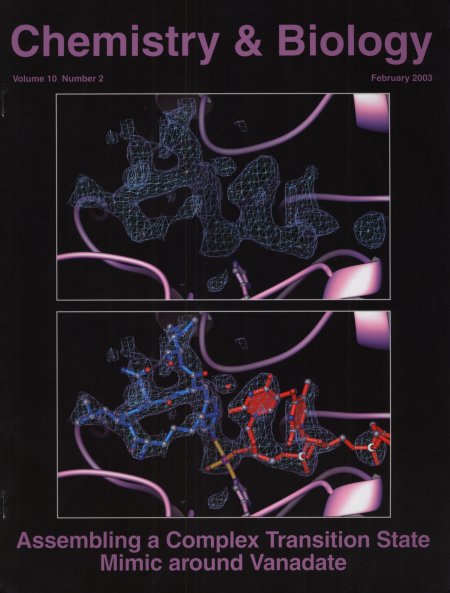
Crystal Structure of a Transition State Mimic for Tyrosyl-DNA
Phosphodiesterase (Tdp1) Assembled from Vanadate,DNA, and a Topoisomerase
I-derived Peptide.
(Doug Davies)
Section I 1.4 Iron-Dependent Repressor IdeR from M. tuberculosis

The crystal structure of cobalt-activated IdeR-(mbtA-mbtB) operator DNA complex. Subunits A,B,C and D of IdeR colored in red, orange, blue, and magenta, respectively. The 33mer DNA duplex containing the mbtA-mbtB operator sequence is shown in a stick model labeled strands E and F for mbtA and mbtB operators, respectively, and is distorted from canonical B-DNA.
Two IdeR dimers bind to two opposite sides of the DNA, forming a double-dimer complex. The CD dimer is shown with its 2-fold in the plane of the paper while the AB dimer does not lie in the same plane.
Each IdeR subunit contains three cobalt ions (pink spheres) and one sodium ion (green spheres). Metal-binding site 1 is located in the dimerization domain with two extra ligands provided by the SH3-like third domain. Metal-binding site 2 is coordinated by residues from the N-terminal DNA-binding domain and the dimerization domain.
Each subunit of IdeR binds to the DNA using the DNA-binding
domain with extensive contacts in the major groove of the DNA by the HTH motif.
(George Wisedchaisri)

Schematic representation of the IdeR-DNA contacts. The protein-DNA
interactions are very similar in the two complexes per asymmetric unit. Amino
acid residues of IdeR contacting the DNA are colored by subunit, red for A,
orange for B, blue for C, and magenta for D. The 19 base-pair "iron box" of
the DNA is highlighted in bright blue where the region outside this box is
in gray. The base-pair G15E-C18F is at the center of the iron box. Protein-DNA
backbone hydrogen bonds (<3.5Å) are shown as black broken arrows. Protein-nucleotide
base interaction (<3.9Å) are highlighted in colored boxes and arrows.
Red boxes represent thymine-specific interactions for Ser37 and Pro39. Gold
boxes represent interactions by Gln43, and green boxes for Pro39 only.
(George Wisedchaisri)
Section I 1.5 Malaria Peptide Deformylase
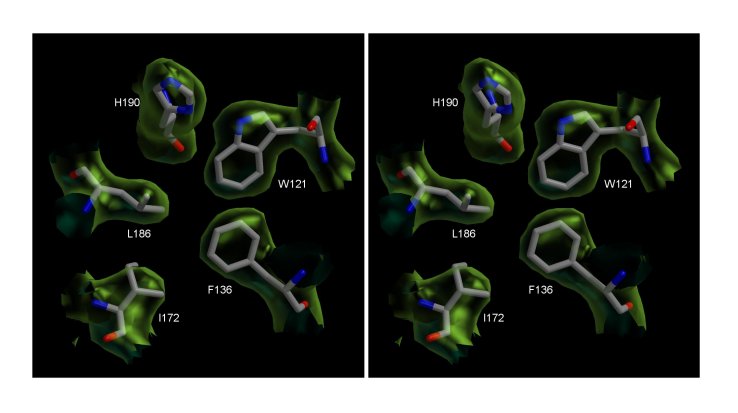
The core of Peptide Deformylase (PDF) from the malaria
parasite Plasmodium falciparum. Stereo view of the σA-Weighted
2Fo-Fo electron density map countoured at 1 σ.
The figure shows a section of the density in the hydrophobic core region of
the structure and includes the final model.
(Abhinav Kumar)
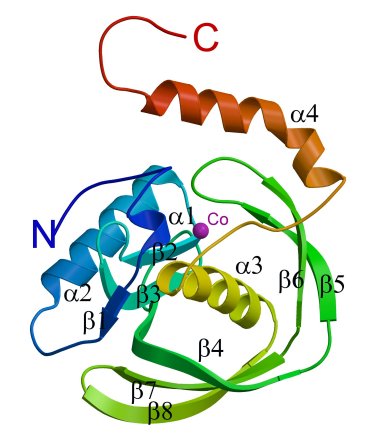
A ribbon diagram showing the secondary structural elements
of the PfPDF subunit.The cobalt position is indicated by a purple sphere.
The kink in the C-terminal helix α4 is quite distinct.
(Abhinav Kumar)
For more information about this subject, please see the Research Summary text, Part I Section 1.